Point Modes vs Cluster Cores¶
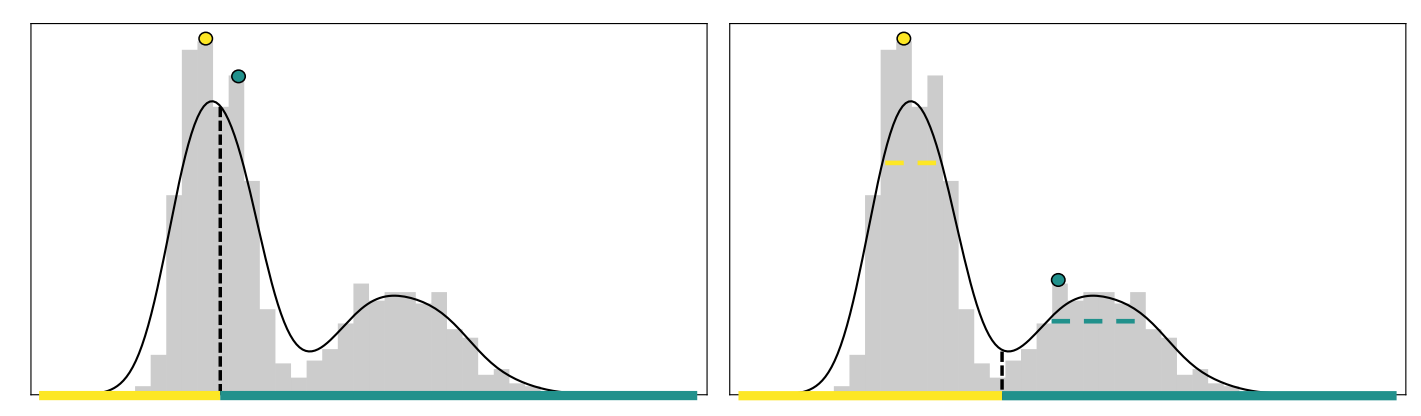
An illustrative example demonstrating the benefits of seeking cluster cores of the density. The black curve represents the underlying density and the grey histogram represents a sample from the density. Left: DPC incorrectly selects both centers from the first cluster, as the noise in the density estimate causes the peak-finding method to favor the high-density cluster. Right: Cluster cores, represented by dashed lines, better represent the cluster centers.
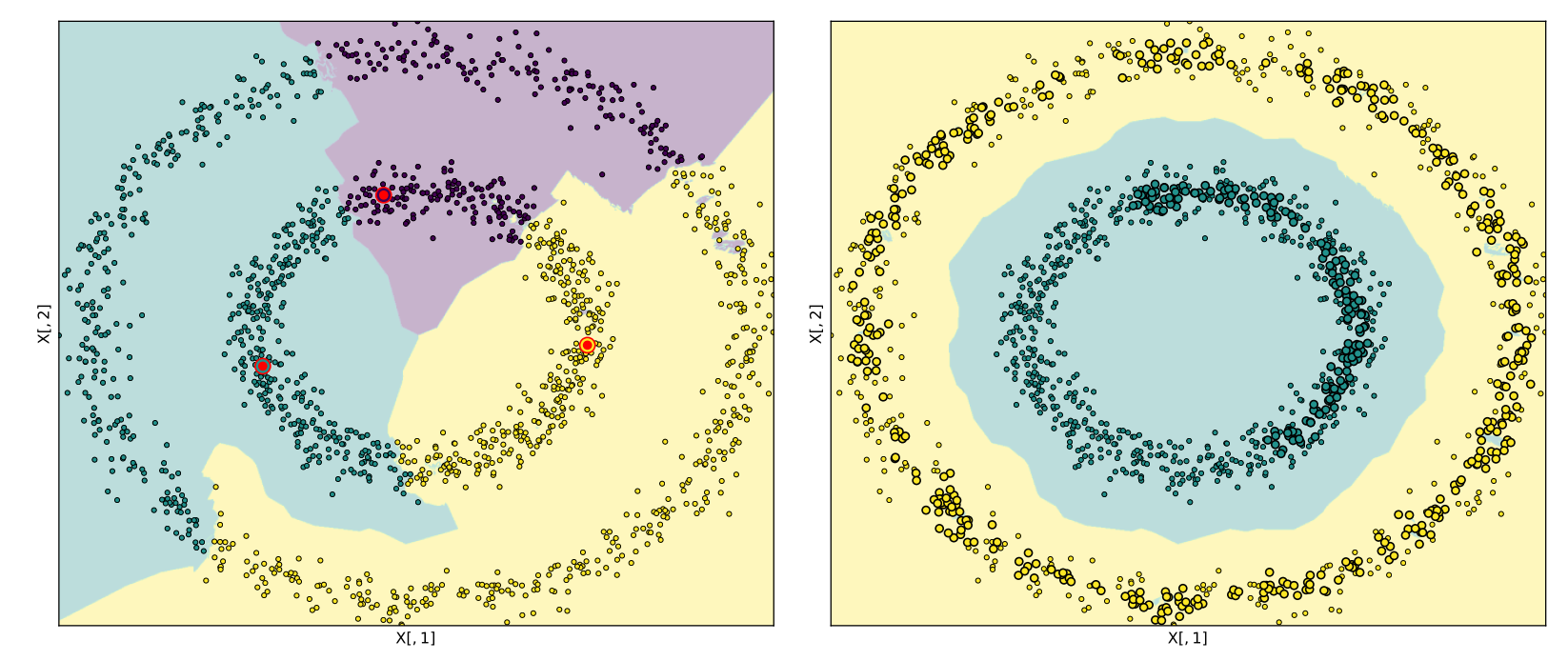
Comparison of the DPC and DCF methods when applied to the Noisy Circles dataset. The Noisy Circles dataset contains two clusters, a high-density cluster (inner circle) and a low-density cluster (outer circle). The DPC method, as seen on the left of the figure, proceeds by searching for the points with maximal values of the peak-finding criterion. This method erroneously selects the multiple centers from the inner cluster. The allocation mechanism incorrectly assigns all points in the outer cluster. For this example, seven points in the inner cluster have larger value of the peak-finding criterion than the maximum value in the outer cluster.
The DCF procedure can be seen on the right of the figure. We first select the instance of maximum density as the first peak. However, DCF proceeds to compute the cluster core associated with this point, the highlighted larger green points, and remove all elements of the core from consideration as centers. Of those remaining, the point with maximal value of the peak-finding criterion is in the outer cluster. The associated cluster core is visible in yellow. As no edge in the $k$-NN graph exists between this cluster core and the first cluster core, it is accepted as a valid cluster core. The algorithm's termination procedure is invoked when assessing a third center. The third center is selected as before; however, as the cluster core associated with this point contains all of the instances in the dataset, the algorithm terminates.